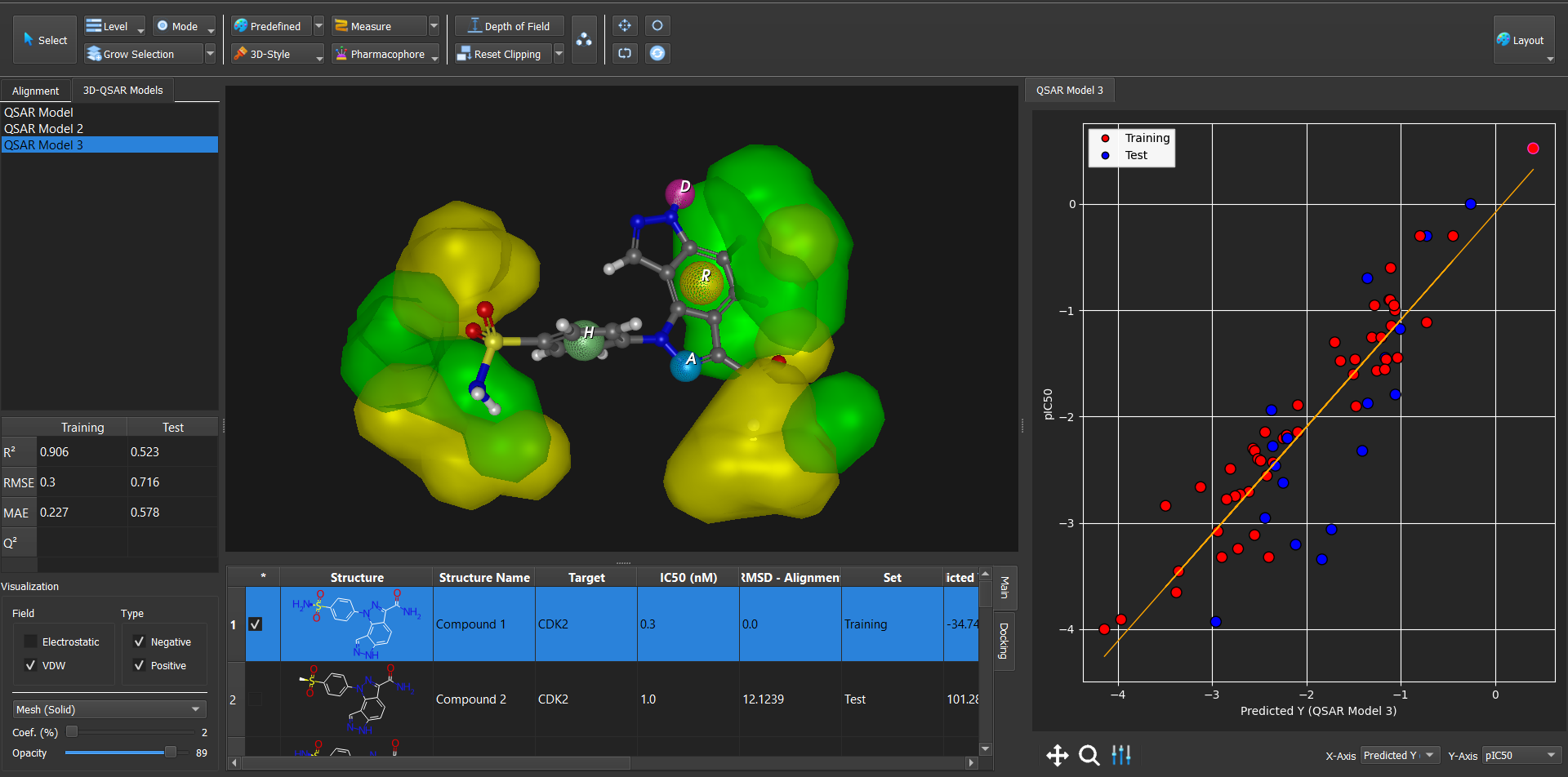
Dockamon
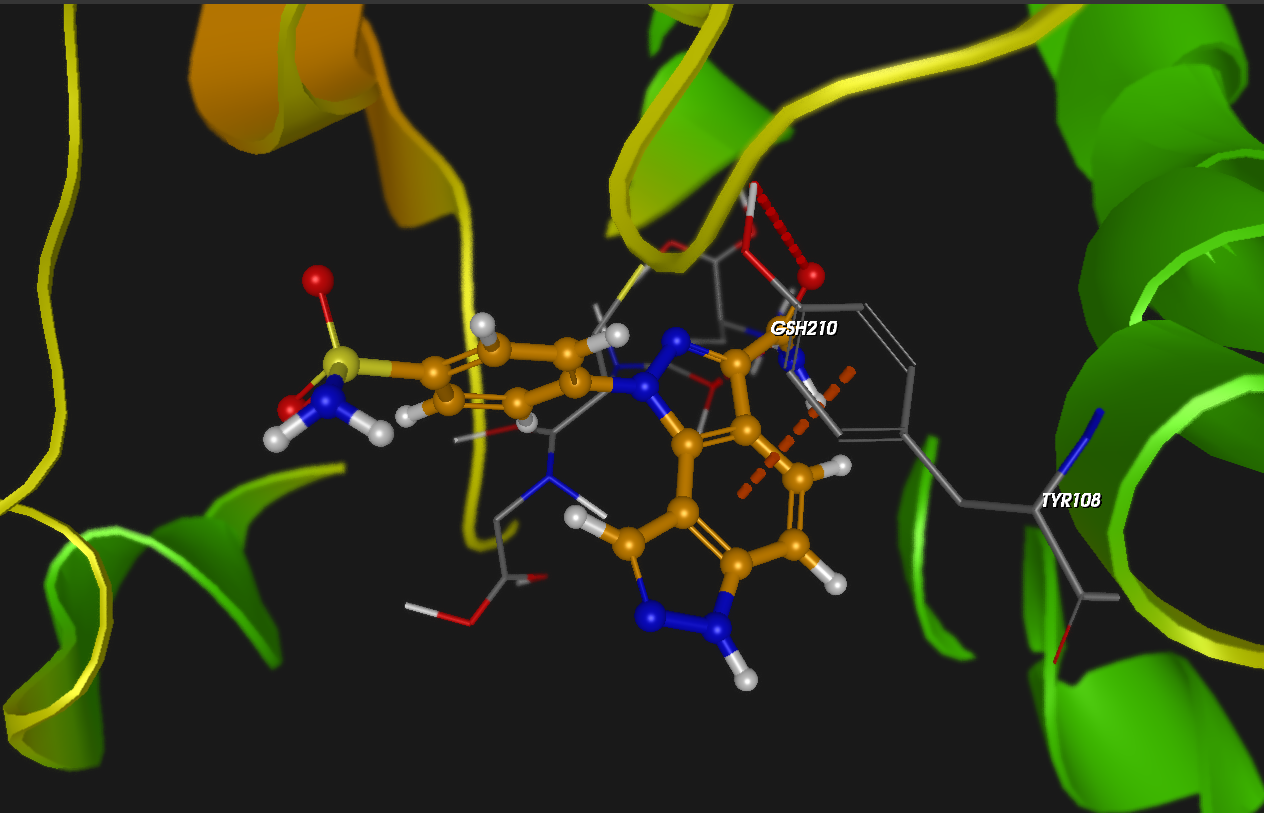
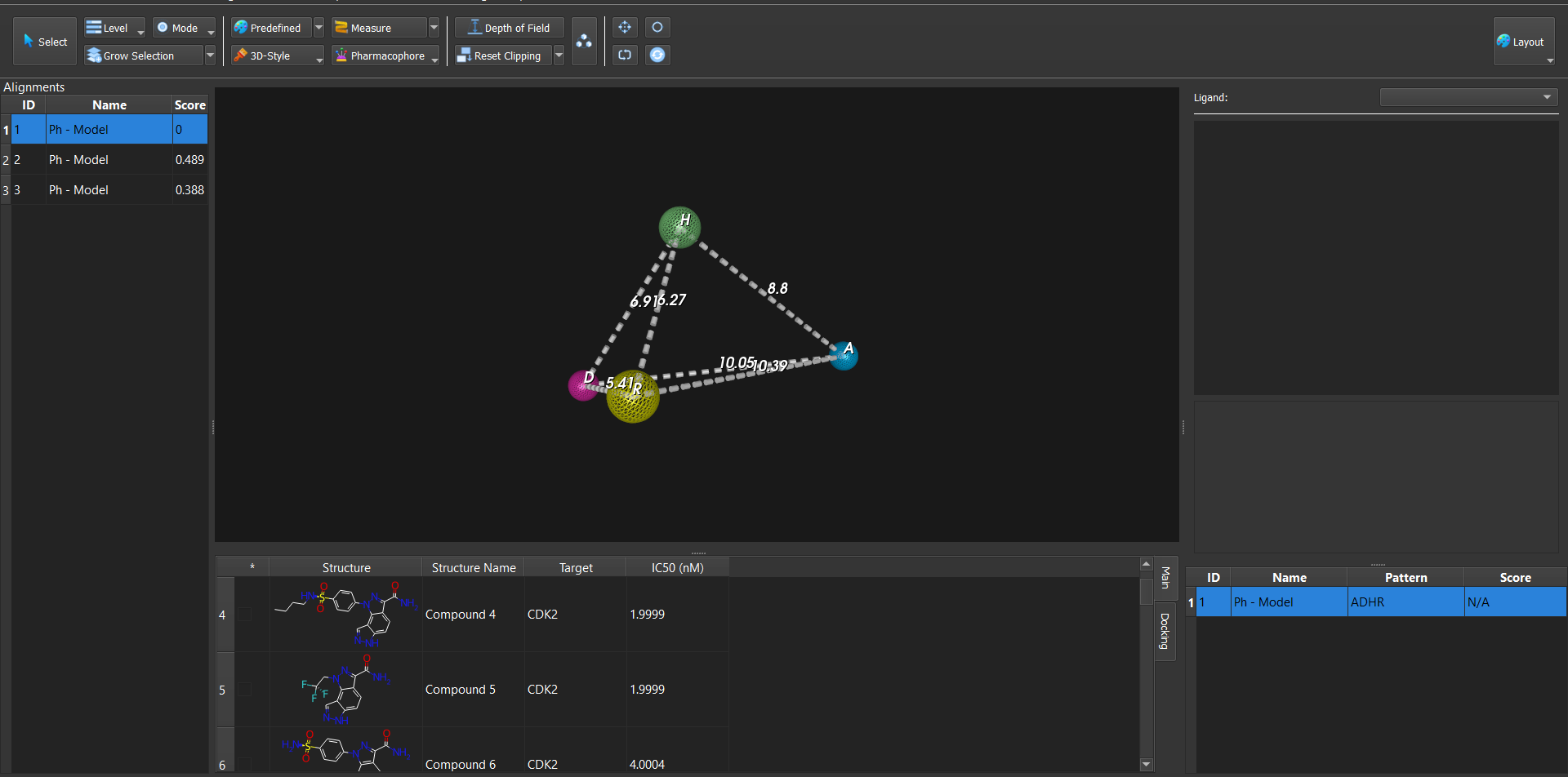
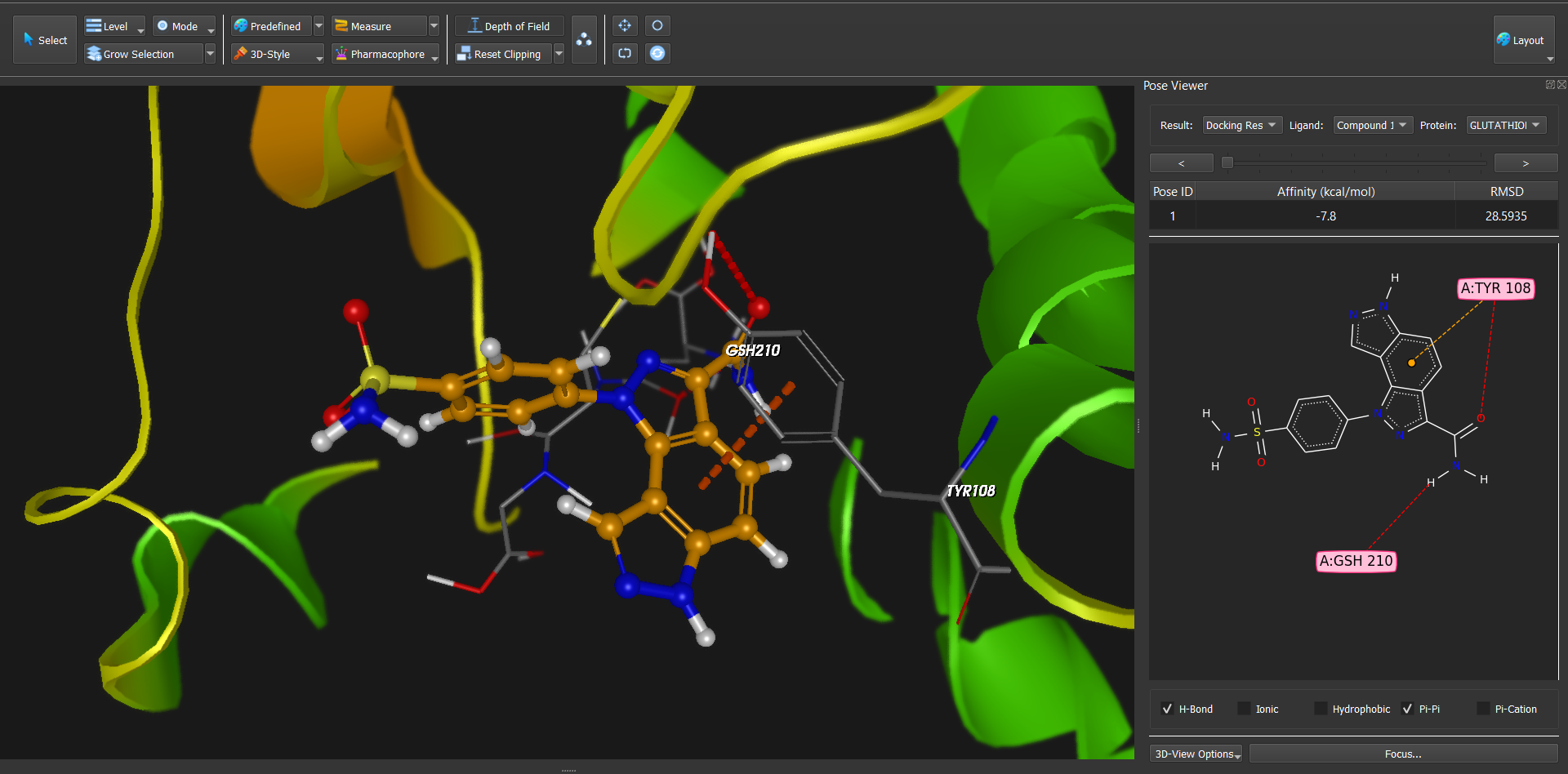
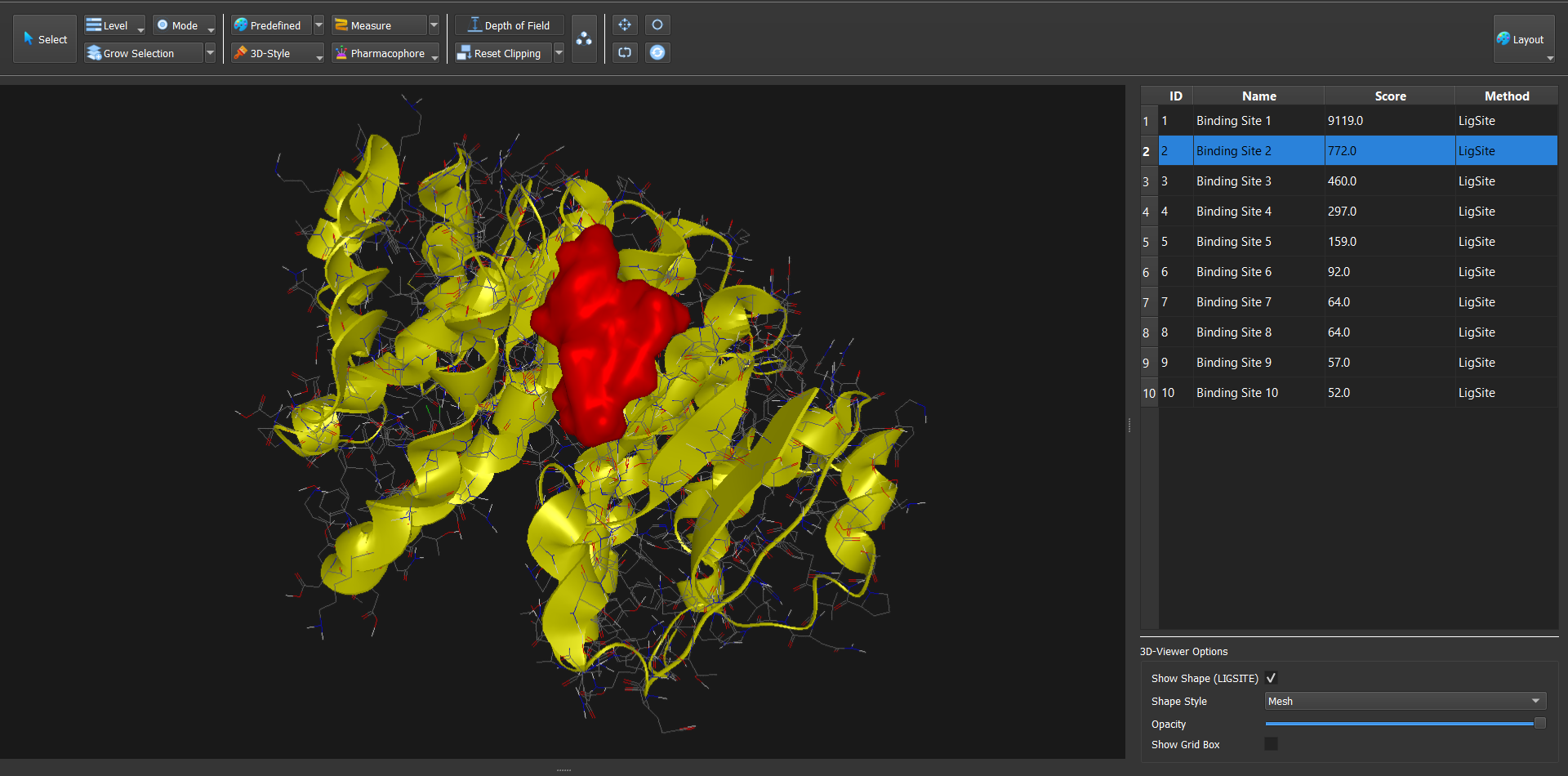
Dockamon is a computer-aided drug design (CADD) software offering pharmacophore modeling, 3D-QSAR, molecular docking, and other advanced tools. It combines comprehensive functionality with a modern, user-friendly interface, integrating both structure-based and ligand-based drug design methods into a single unified platform.