PyRx
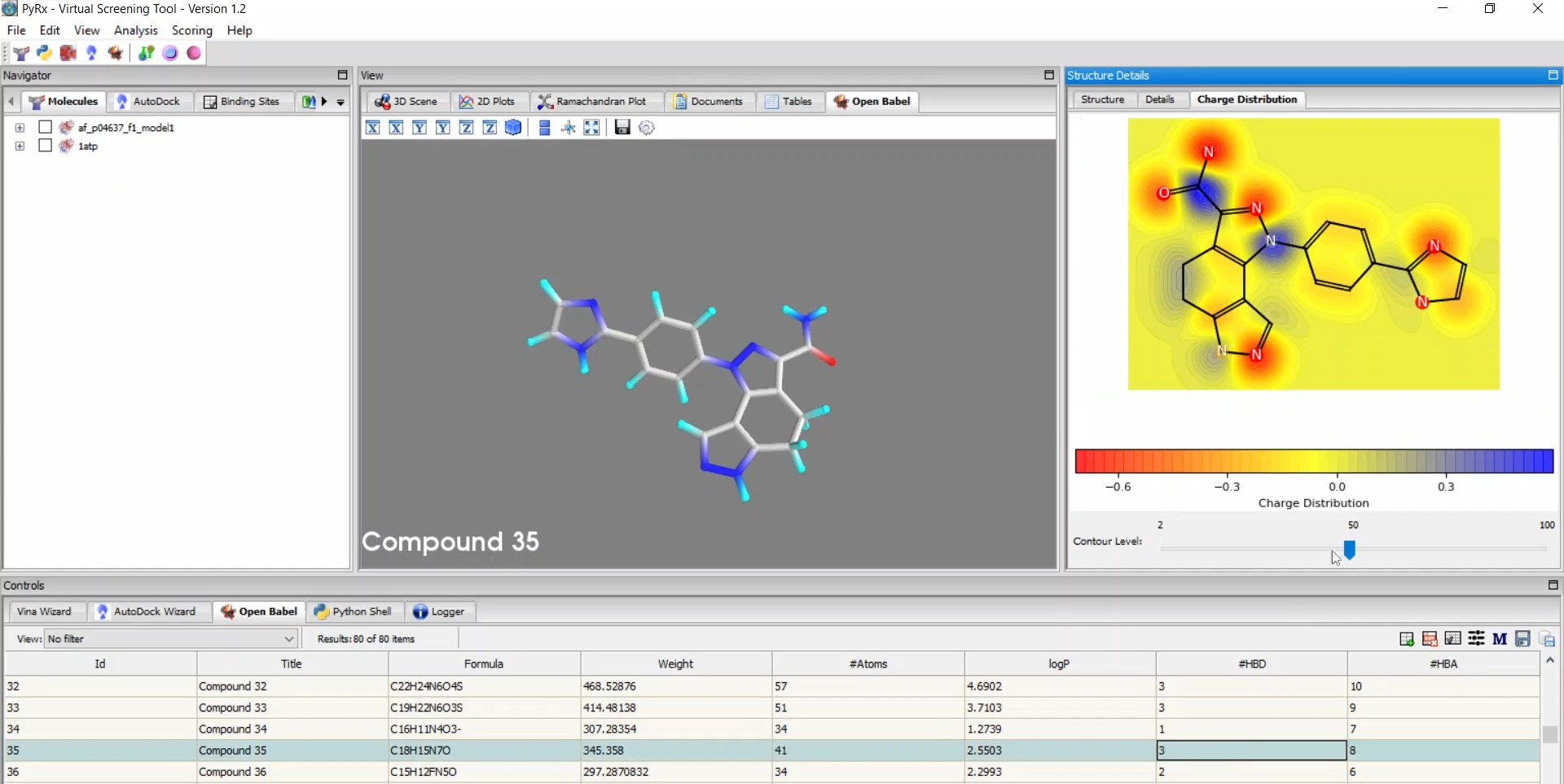
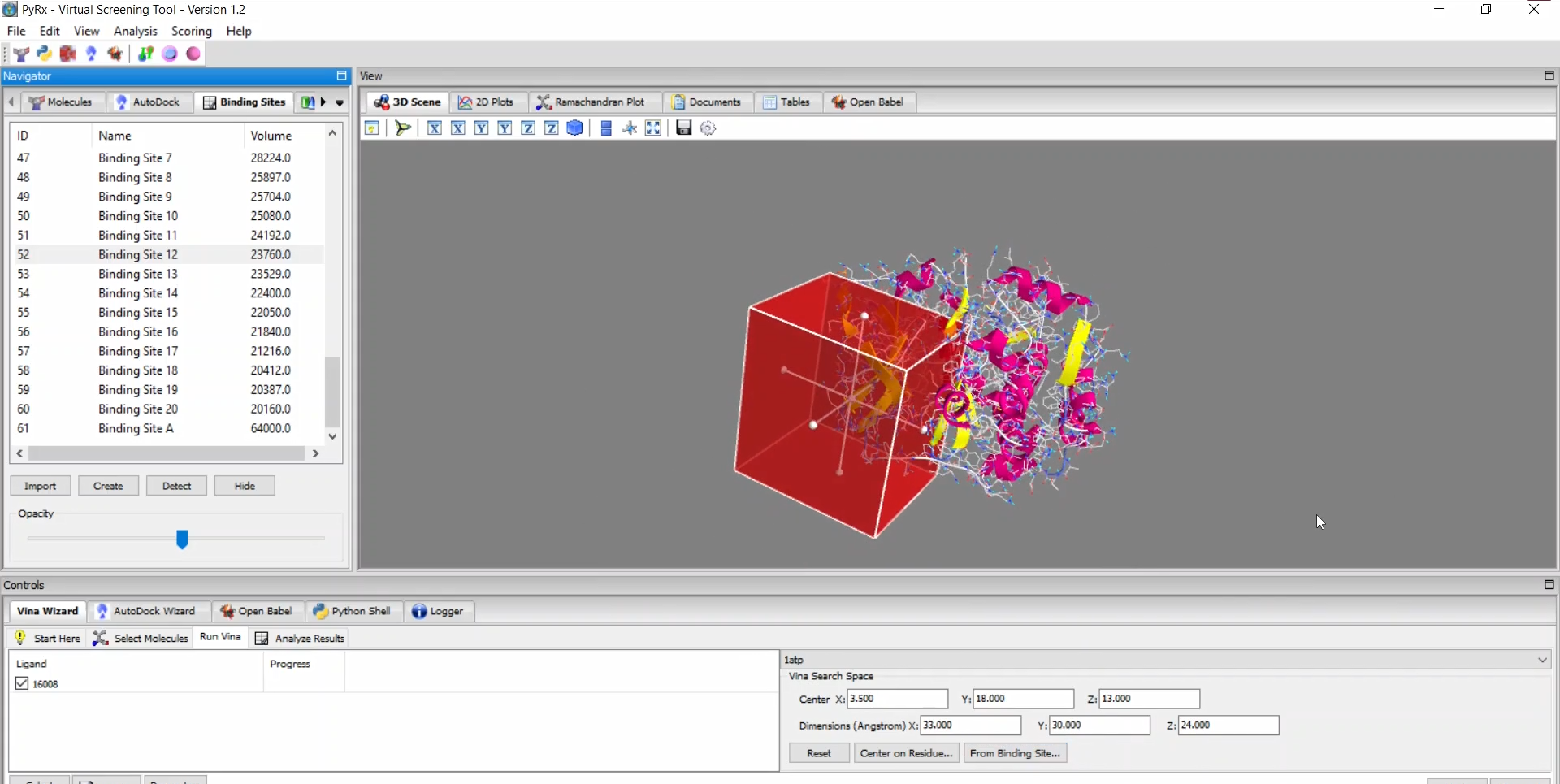
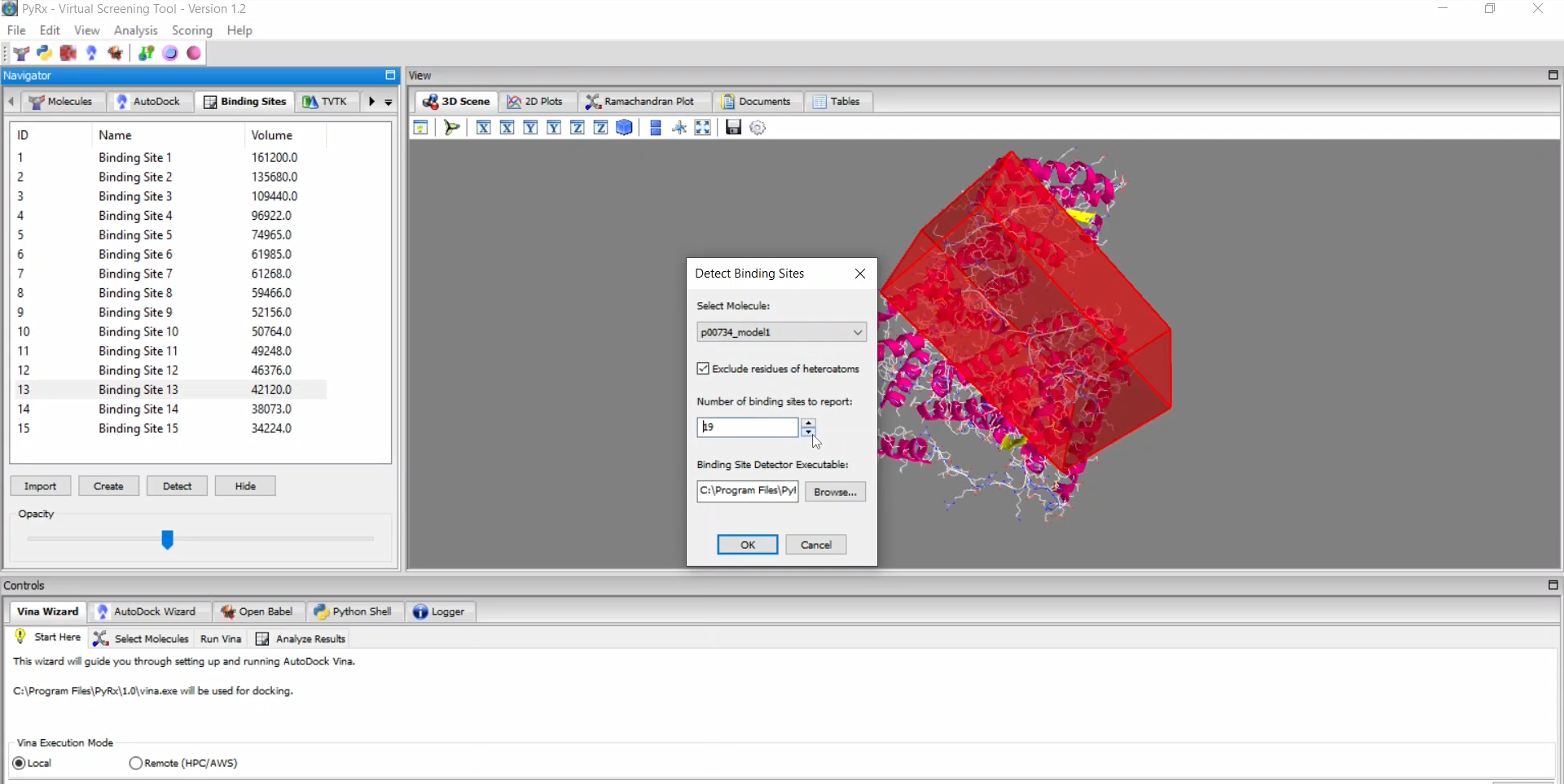
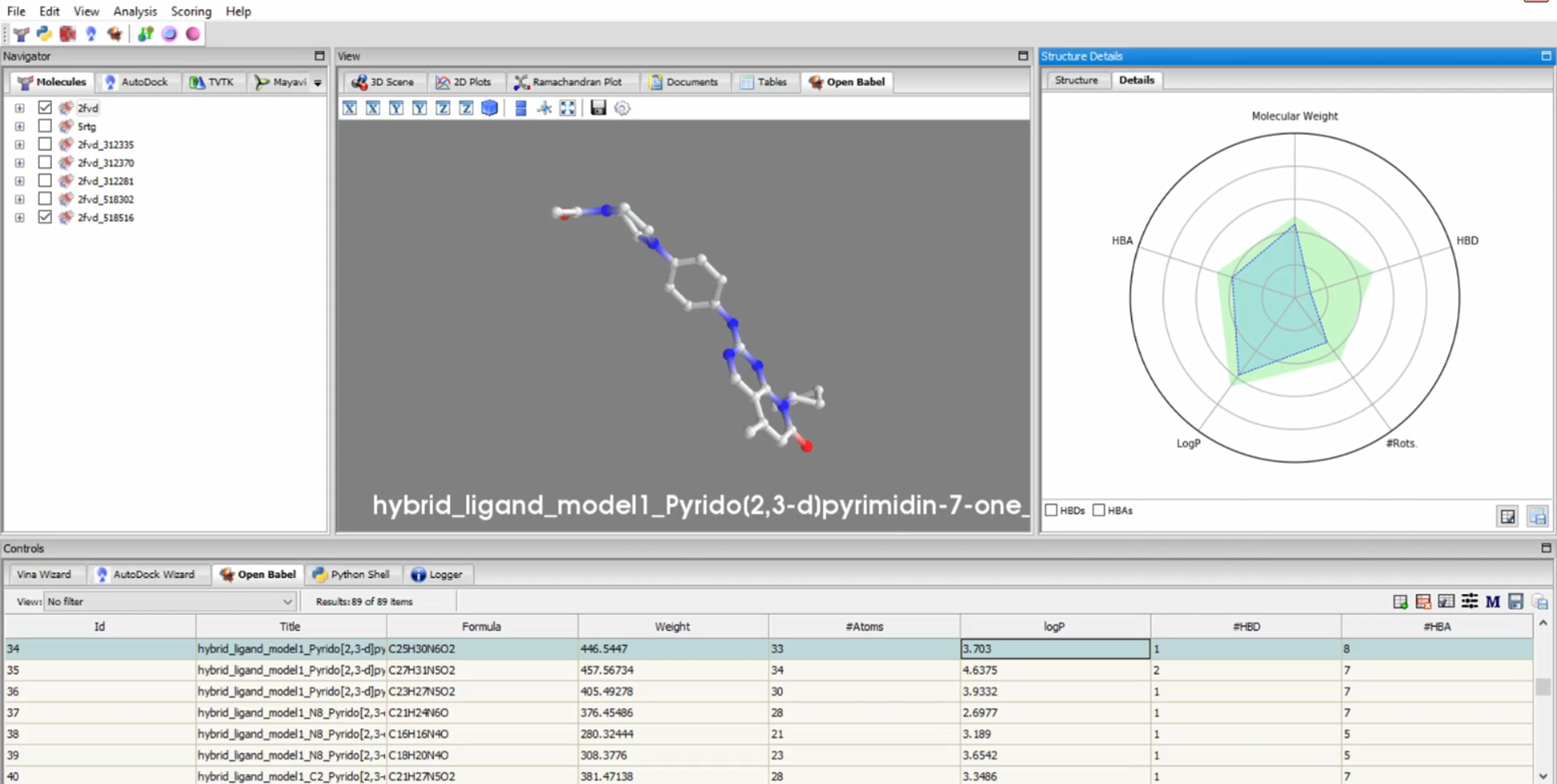
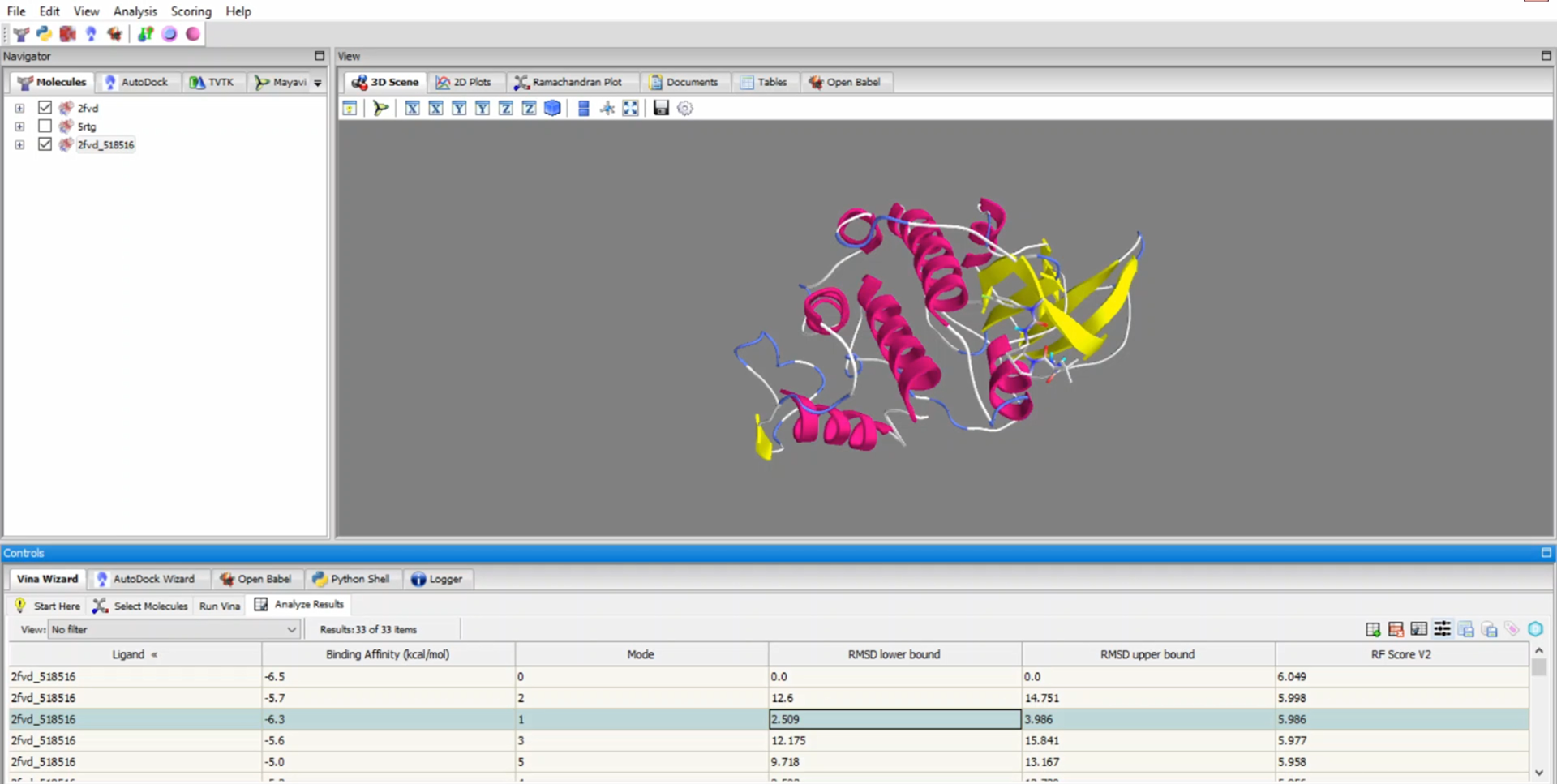
PyRx is a structure-based drug design software primarily used for virtual screening through molecular docking. As one of the most popular and highly cited tools in drug discovery and bioinformatics, PyRx enables researchers to easily screen large compound libraries against target proteins. Originally developed by Sarkis Dallakyan, PyRx has seen continued enhancements in recent years with CrescentSilico contributing to the development of new features.
Official page: https://pyrx.sourceforge.io/